-
2024
- Xiaolin Liang, Meng Gong, Zhikai Wang, Jie Wang, Weiwei Guo, Aoling Cai, Zhenye Yang, Xing Liu, Fuqiang Xu, Wei Xiong, Fu C, Xiangting Wang, “LncRNA TubAR complexes with TUBB4A and TUBA1A to promote microtubule assembly and maintain myelination”, Cell Discov,2024 May 21;10(1):54. doi: 10.1038/s41421-024-00667-y.
- Shi-Yan Sun, Lingyun Nie, Jing Zhang, Xue Fang, Hongmei Luo, Fu C, Zhiyi Wei, Ai-Hui Tang, “The interaction between KIF21A and KANK1 regulates dendritic morphology and synapse plasticity in neurons”, Neural Regen Res,2025 Jan 1;20(1):209-223. doi: 10.4103/1673-5374.391301. Epub 2023 Dec 21.
- Ling Liu, Yifan Wu, Ke Liu, Mengdan Zhu, Shouhong Guang, Fengsong Wang, Xing Liu, Xuebiao Yao, Jiajia He, Fu C, “The absence of the ribosomal protein Rpl2702 elicits the MAPK-mTOR signaling to modulate mitochondrial morphology and functions”, Redox Biol,2024 Apr 29:73:103174. doi: 10.1016/j.redox.2024.103174.
- Shengnan Zheng, Biyu Zheng, Fu C, “The Roles of Septins in Regulating Fission Yeast Cytokinesis”, J Fungi (Basel), 2024 Jan 30;10(2):115. doi: 10.3390/jof10020115.
- Zhen Dou, Ran Liu, Ping Gui, Fu C, Jennifer Lippincott-Schwartz, Xuebiao Yao, Xing Liu, “Fluorescence complementation-based FRET imaging reveals centromere assembly dynamics”, Mol Biol Cell, 2024 Apr 1;35(4):ar51. doi: 10.1091/mbc.E23-09-0379. Epub 2024 Feb 21.
- Mengdan Zhu, Zheng Fang, Yifan Wu, Fenfen Dong, Yuzhou Wang, Fan Zheng, Xiaopeng Ma, Shisong Ma, Jiajia He, Xing Liu, Xuebiao Yao, Fu C, “A KDELR-mediated ER-retrieval system modulates mitochondrial functions via the unfolded protein response in fission yeast”, J Biol Chem, 2024 Mar;300(3):105754. doi: 10.1016/j.jbc.2024.105754.
- Fengrui Yang, Mingrui Ding, Xiaoyu Song, Fang Chen, Tongtong Yang, Chunyue Wang, Chengcheng Hu, Qing Hu, Yihan Yao, Shihao Du, Phil Y Yao, Peng Xia, Gregory Adams Jr, Fu C,Shengqi Xiang, Dan Liu, Zhikai Wang, Kai Yuan, Xing Liu, “Organization of microtubule plus-end dynamics by phase separation in mitosis.”, J Mol Cell Biol, 2024 Feb 6:mjae006. doi: 10.1093/jmcb/mjae006. Online ahead of print.
- He J, Liu K, Fu C, “Recent insights into the control of mitochondrial fission.”, Biochem Soc Trans, January 30, 2024. doi:10.1042/BST20230220
- Wei Liu, Zhen Dou, Chunyue Wang, Gangyin Zhao, Fengge Wu, Chunli Wang, Felix Aikhionbare, Mingliang Ye, Divine Mensah Sedzro, Zhenye Yang, Fu C, Zhikai Wang, Xinjiao Gao, Xuebiao Yao, Xiaoyu Song, Xing Liu, “Aurora B promotes the CENP-T-CENP-W interaction to guide accurate chromosome segregation in mitosis.”, J Mol Cell Biol, 2024 Jan 10:mjae001. doi: 10.1093/jmcb/mjae001. Online ahead of print.
- Jian Y, Jiang Y, Nie L, Dou Z, Liu X, Fu C, “Phosphorylation of Bub1 by Mph1 promotes Bub1 signaling at the kinetochore to ensure accurate chromosome segregation.”, J Biol Chem, 2023 Dec 13;doi:10.1016/j.jbc.2023.105559
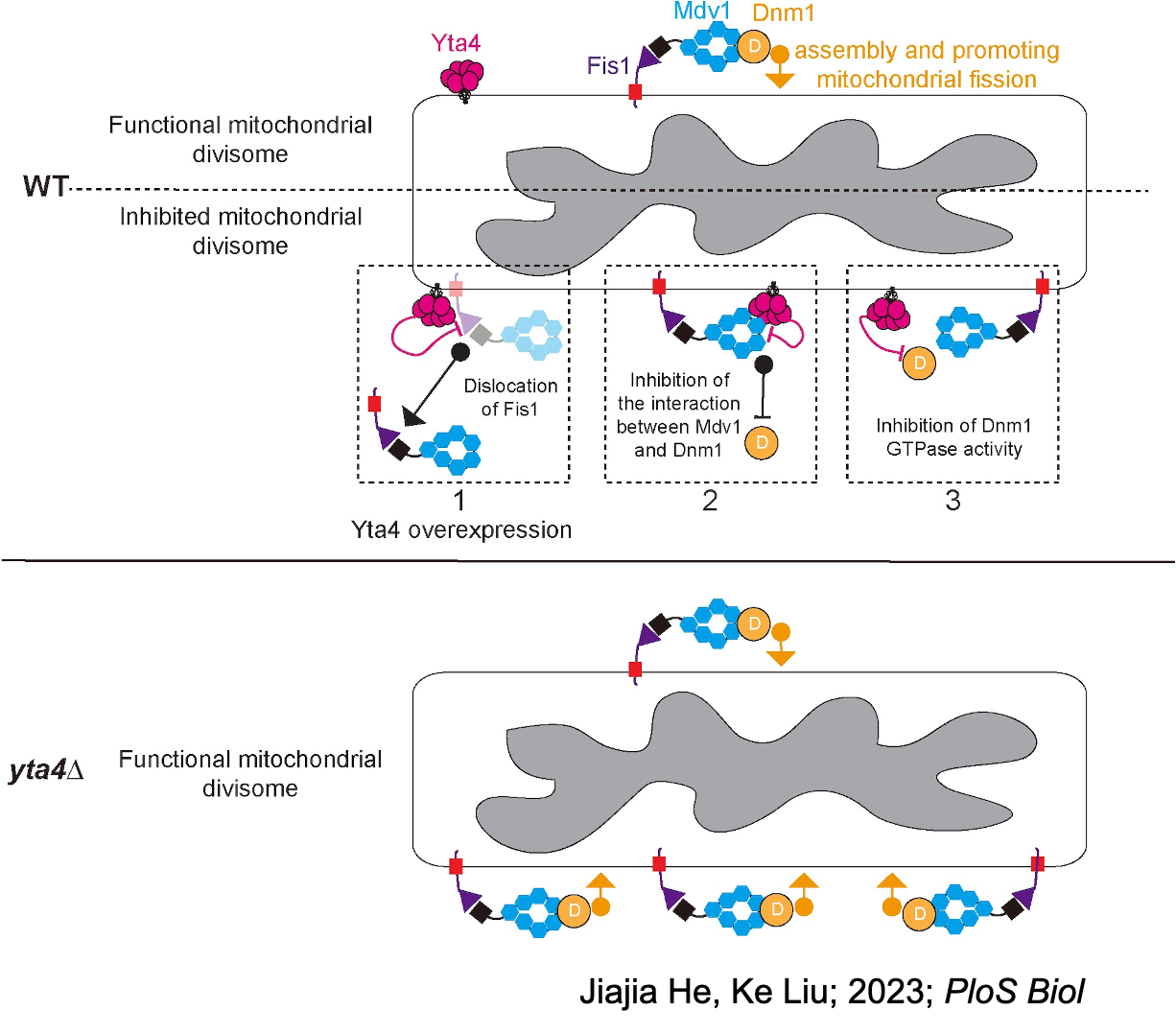
- He J, Liu K, Wu Y, Zhao C, Yan S, Chen JH, Hu L, Wang D, Zheng F, Wei W, Xu C, Huang C, Liu X, Yao X, Ding L, Fang Z, Tang AH, Fu C, “The AAA-ATPase Yta4/ATAD1 interacts with the mitochondrial divisome to inhibit mitochondrial fission”, PLoS Biol, 2023 Aug 17;21(8):e3002247. doi: 10.1371/journal.pbio.3002247. Epub ahead of print. PMID: 37590302.
- Xu D, Chen X, Kuang Y, Hong M, Xu T, Wang K, Huang X,Fu C,Ruan K, Zhu C, Feng X, Guang S. “ rRNA intermediates coordinate the formation of nucleolar vacuoles in C. elegans”, Cell Rep,2023 Aug 1;42(8):112915. doi: 10.1016/j.celrep.2023.112915. Epub ahead of print. PMID: 37537842.
- Liu R, Dou Z, Tian T, Gao X, Chen L, Yuan X, Wang C, Hao J, Gui P, Mullen M, Aikhionbare F, Niu L, Bi G, Zou P, Zhang X, Fu C,Yao X, Zang J, Liu X. “Dynamic phosphorylation of CENP-N by CDK1 guides accurate chromosome segregation in mitosis”, J Mol Cell Biol, 2023 Jun 26:mjad041. doi: 10.1093/jmcb/mjad041. Epub ahead of print. PMID: 37365681.
- Li J, Yuan X, Yang F, Cao J, Akram S, Zou P, Aikhionbare F, Li X, Chen Y, Fu C,Wang Z, Liu X, Yao X. “PML-LRIF1 interactions form a novel link between PML bodies and centromeres”, J Mol Cell Biol, 2023 Jun 7:mjad038. doi: 10.1093/jmcb/mjad038. Epub ahead of print. PMID: 37286714.
- Chen Z, Zheng S, Fu C, “Shotgun knockdown of RNA by CRISPR-Cas13d in fission yeast”, J Cell Sci, 2023 Mar 15;136(6):jcs260769. doi: 10.1242/jcs.260769. Epub 2023 Mar 20. PMID: 36825467.
- Xu T, Liao S, Huang M, Zhu C, Huang X, Jin Q, Xu D, Fu C,Chen X, Feng X, Guang S. “A ZTF-7/RPS-2 complex mediates the cold-warm response in C. elegans”, PLoS Genet, 2023 Feb 10;19(2):e1010628. doi: 10.1371/journal.pgen.1010628. PMID: 36763670; PMCID: PMC9949642.
- Wu D, Chu Y, Wei W, Liu L, Fu C, “Fission yeast cells mix parental mitochondria in a progressive manner during meiosis”, J Mol Cell Biol, 2022 Dec 16; mjac070. doi: 10.1093/jmcb/mjac070
- Jian Y, Nie L, Liu S, Jiang Y, Dou Z, Liu X, Yao X*, Fu C*, “Fission yeast CENP-S/X regulates the spindle assembly checkpoint and faithful chromosome segregation”, J Cell Sci, 2022.(*co-corresponding authors) doi: 10.1242/jcs.260124. 2022 Dec 20.
- Wei W, Zheng B, Zheng S, Wu D, Chu Y, Zhang S, Wang D, Ma X, Liu X, Yao X*, Fu C*, “The Cdc42 GAP Rga6 promotes monopolar outgrowth of spores”, J Cell Biol, 2022.(*co-corresponding authors) doi: 10.1083/jcb.202202064. Epub 2022 Nov 10.
- Wu Z, Xu H, Wang P, Liu L, Cai J, Chen Y, Zhao X, You X, Liu J, Guo X, Xie T, Feng J, Zhou F, Li R, Xie Z, Xue Y*, Fu C*, Liang Y* “The entry of unclosed autophagosomes into vacuoles and its physiological relevance”, PLoS Genet, 2022.(*co-corresponding authors) doi: 10.1371/journal.pgen.1010431. 2022 Oct 13;18(10):e1010431.
- Wang X, Zheng F, Yi YY, Wang GY, Hong LX, McCollum D, Fu C*,Wang Y*, Jin QW.* “Ubiquitination of CLIP-170 family protein restrains polarized growth upon DNA replication stress”, Nature Commun, 2022.(*co-corresponding authors) doi: 10.1038/s41467-022-33311-y. 2022 Sep 22;13(1):5565.
- Ge X, He Z, Cao C, Xue T, Jing J, Ma R, Zhao W, Liu L, Jueraitetibaike K, Ma J, Feng Y, Qian Z, Zou Z, Chen L, Fu C*, Song N*, Yao B* “Protein palmitoylation-mediated palmitic acid sensing causes blood-testis barrier damage via inducing ER stress”, Redox Biol, 2022.(*co-corresponding authors) doi: 10.1016/j.redox.2022.102380. Epub 2022 Jul 2
- He J, Liu K, Zheng S, Wu Y, Zhao C, Yan S, Liu L, Ruan K, Ma X*, Fu C* “The acyl-CoA-binding protein Acb1 regulates mitochondria, lipid droplets, and cell proliferation”, FEBS Lett, 2022.(*corresponding authors) doi: 10.1002/1873-3468.14415. 2022 Jul;596(14):1795-1808
- Shen S, Jian Y, Cai Z, Li F, Lv M, Liu Y, Wu J*, Fu C*, Shi Y* “Structural insights reveal the specific recognition of meiRNA by the Mei2 protein”, J Mol Cell Biol, 2022.(*co-corresponding authors) doi: 10.1093/jmcb/mjac029. 2022 May 5;mjac029
- Liu S, Yuan X, Gui P, Liu R, Durojaye O, Hill DL , Fu C, Yao X, Dou Z, Liu X “Mad2 promotes Cyclin B2 recruitment to the kinetochore for guiding accurate mitotic checkpoint”, EMBO Rep, 2022. doi: 10.15252/embr.202154171. Epub 2022 Apr 5
- Sheng X, Liu C, Yan G, Li G, Liu J, Yang Y, Li S, Li Z, Zhou J, Zhen X, Zhang Y, Diao Z, Hu Y, Fu C, Yao B, Li C, Cao Y, Lu B, Yang Z, Qin Y, Sun H, Ding L “The mitochondrial protease LONP1 maintains oocyte development and survival by suppressing nuclear translocation of AIFM1 in mammals”, EBioMedicine, 2022. doi: 10.1016/j.ebiom.2021.103790.Epub 2021 Dec 30.
- Zheng S, Zheng B, Liu Z, Liu X, Yao X, Wei W* Fu C* “The Cdc42 GTPase activating protein Rga6 promotes the cortical localization of Septin”, J Cell Sci, 2022.(*corresponding authors) doi: 10.1242/jcs.259228. Epub 2022 Feb 16
- Fenfen Dong, Mengdan Zhu, Fan Zheng, Fu C*“Mitochondrial fusion and fission are required for proper mitochondrial function and cell proliferation in fission yeast.”, FEBS J(Cover story), 2022.(*corresponding authors) doi: 10.1111/febs.16138. Epub 2021 Aug 3
- Rasul F, Zheng F, Dong F, He J, Liu L, Liu W, Cheema J, Wei W*, Fu C* “Emr1 regulates the number of foci of the endoplasmic reticulum-mitochondria encounter structure complex.”, Nature Commun, 2021.(*corresponding authors) doi: 10.1038/s41467-020-20866-x
- Hu C , Wang X , Liu L , Fu C, Chu K , Smith ZJ “Fast confocal Raman imaging via context-aware compressive sensing.”, Analyst, 2021. doi: 10.1039/d1an00088h. Epub 2021 Feb 24
- Wang X, Wang W, Wang X, Wang M, Zhu L, Garba F, Fu C, Zieger B, Liu X, Liu X, Yao X “Fast confocal Raman imaging via context-aware compressive sensing.”, J Mol Cell Biol, 2021. doi: 10.1093/jmcb/mjab036. 2021 Sep 11;13(6):395-408
- Zheng F, Dong F, Yu S, Li T, Jian Y, Nie L, Fu C*“Klp2 and Ase1 synergize to maintain meiotic spindle stability during metaphase I.”, J Biol Chem, 2020.(*corresponding authors) doi: 10.1074/jbc.RA120.012905
- Zhang M, Zheng F, Xiong Y, Shao C, Wang C, Wu M, Niu X, Dong F, Zhang X*, Fu C*, Zang J* “Centromere targeting of Mis18 requires the interaction with DNA and H2A-H2B in fission yeast.”, Cell Mol Life Sci, 2021.( *co-corresponding authors) doi: 10.1007/s00018-020-03502-1
- Zheng F, Jia B, Dong F, Liu L, Rasul F, He J, Fu C*“Glucose starvation induces mitochondrial fragmentation depending on the dynamin GTPase Dnm1/Drp1 in fission yeast.”, J Biol Chem, 2019.(*corresponding authors) doi: 10.1074/jbc.RA119.010185. Epub 2019 Sep 27.
- Liu W, Zheng F, Wang Y, Fu C*“ Alp7-Mto1 and Alp14 synergize to promote interphase microtubule regrowth from the nuclear envelope.”, J Mol Cell Biol, 2019.(*corresponding authors) doi: 10.1093/jmcb/mjz038
- Niu X, Zheng F*, Fu C*“The concerted actions of Tip1/CLIP-170, Klp5/Kinesin-8, and Alp14/XMAP215 regulate microtubule catastrophe at the cell end”, J Mol Cell Biol, 2019.(*corresponding authors) doi: 10.1093/jmcb/mjz039. 2019 Dec 23;11(11):956-966
- Shen J, Li T, Niu X, Liu W, Zheng S, Wang J, Wang F, Cao X, Yao X, Zheng F*, Fu C*, “The J-domain cochaperone Rsp1 interacts with Mto1 to organize noncentrosomal microtubule assembly”, Mol Biol Cell, 2019.(*corresponding authors). doi: 10.1091/mbc.E18-05-0279. 2019 Jan 15;30(2):256-267
- Liu G, Dong F, Fu C*,Smith ZJ* “Automated morphometry toolbox for analysis of microscopic model organisms using simple bright-field imaging”, Biol Open, 2019.(*co-corresponding authors). doi: 10.1242/bio.037788. 2019 Mar 12;8(3):bio037788
- Ding M, Jiang J, Yang F, Zheng F, Fang J, Wang Q, Wang J, Yao W, Liu X, Gao X, Mullen M, He P, Rono C, Ding X, Hong J, Fu C, Liu X, Yao X “Holliday junction recognition protein interacts with and specifies the centromeric assembly of CENP-T”, J Biol Chem, 2019. doi: 10.1074/jbc.RA118.004688. 2019 Jan 18;294(3):968-980
- Zheng S, Dong F, Rasul F, Yao X, Jin Q, Zheng F*, Fu C* “Septins regulate the equatorial dynamics of the separation initiation network kinase Sid2p and glucan synthases to ensure proper cytokinesis”, FEBS J, 2018.(*co-corresponding authors). doi: 10.1111/febs.14487. Epub 2018 May 14
- Wang L, Li S, Sun Z, Wen G, Zheng F, Fu C, Li H “Segmentation of yeast cell's bright-field image with an edge-tracing algorithm”, J Biomed Opt, 2018. doi: 10.1117/1.JBO.23.11.116503. 2018 Nov;23(11):1-7
- Zhou X, Zheng F, Wang C, Wu M, Zhang X, Wang Q, Yao X, Fu C, Zhang X, Zang J“Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis.”, Proc Natl Acad Sci U S A, 2017. doi: 10.1073/pnas.1710506114. Epub 2017 Nov 27
- Qin B, Cao D, Wu H, Mo F, Shao H, Chu J, Powell M, Aikhionbare F, Wang D, Fu C, He P, Pan W, Wang W, Liu X, Yao X“ Phosphorylation of SKAP by GSK3β ensures chromosome segregation by a temporal inhibition of Kif2b activity.”, Sci Rep, 2016. doi: 10.1038/srep38791. 2016 Dec 16;6:38791
- Zhu Q, Zheng F, Liu AP, Qian J, Fu C*, Lin Y*“Shape Transformation of the Nuclear Envelope during Closed Mitosis.”, Biophys J, 2016.(*co-corresponding authors) doi: 10.1016/j.bpj.2016.10.004
- Hui Z, Zheng F, Lin Y*, Fu C*“The linear and rotational motions of the fission yeast nucleus are governed by the stochastic dynamics of spatially distributed microtubules.”, J Biomech, 2016.(*co-corresponding authors) doi: 10.1016/j.jbiomech.2016.02.017. 2016 May 3;49(7):1034-1041
- Mo F, Zhuang X, Liu X, Yao PY, Qin B, Su Z, Zang J, Wang Z, Zhang J, Dou Z, Tian C, Teng M, Niu L, Hill DL, Fang G, Ding X, Fu C, Yao X“Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation.”, Nat Chem Biol, 2016. doi: 10.1038/nchembio.2017. 2016 Apr;12(4):226-32
- Dou Z, Liu X, Wang W, Zhu T, Wang X, Xu L, Abrieu A, Fu C, Hill DL, Yao X“Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment.”, PNAS, 2014. doi: 10.1073/pnas.1508791112. Epub 2015 Aug 3
- Li T, Zheng F, Cheung M, Wang F Fu C*“Fission yeast mitochondria are distributed by dynamic microtubules in a motor-independent manner.”, Sci Rep, 2014.(corresponding authors) doi: 10.1038/srep11023. 2015 Jun 5;5:11023
- Zheng F, Li T, Jin D, Syrovatkina V, Scheffler K, Tran PT*, Fu C* “Csi1p recruits alp7p/TACC to the SPB for bipolar spindle formation.”, Mol Biol Cell, 2014.(*corresponding authors) doi: 10.1091/mbc.E14-03-0786. Epub 2014 Jul 23
- Wang X, Yu H, Xu L, Zhu T, Zheng F, Fu C, Wang Z, Dou Z“Dynamic autophosphorylation of Mps1 kinase is required for faithful mitotic progression.”, PLOS ONE, 2014. doi: 10.1371/journal.pone.0104723. 2014 Sep 29;9(9):e104723
- Lan J, Zhu Y, Xu L, Yu H, Yu J, Liu X, Fu C, Wang X, Ke Y, Huang H, Dou Z“The 68-kDa telomeric repeat binding factor 1 (TRF1)-associated protein (TAP68) interacts with and recruits TRF1 to the spindle pole during mitosis.”, J Biol Chem, 2014. doi: 10.1074/jbc.M113.526244. 2014 May 16;289(20):14145-56
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, Fang J, Zang J, Cheng J, Teng M, Ding X, Yao X“Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP).”, J Biol Chem, 2014. doi: 10.1074/jbc.M113.529958. 2014 Mar 21;289(12):8326-36
- Zheng F, Li T, Cheung M, Syrovatkina V, Fu C* “Mcp1p tracks microtubule plus ends to destabilize microtubules at cell tips.”, FEBS Lett, 2014.(*corresponding authors) doi: 10.1016/j.febslet.2014.01.055. 2014 Mar 18;588(6):859-65 2013
- Syrovatkina V#, Fu C#, Tran PT“Antagonistic spindle motors and MAPs regulate metaphase spindle length and chromosome segregation.”, Curr Biol,2013.(#equal contributions) doi: 10.1016/j.cub.2013.10.023. Epub 2013 Nov 14
- Zhu T, Dou Z, Qin B, Jin C, Wang X, Xu L, Wang Z, Zhu L, Liu F, Gao X, Ke Y, Wang Z, Aikhionbare F, Fu C, Ding X, Yao X“ Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore.”, J Biol Chem, 2013. doi: 10.1074/jbc.M113.507970. 2013 Dec 13;288(50):36149-59
- Fu C*, Jain D, Costa J, Velve-Casquillas G, Tran PT* “Mmb1p binds mitochondria to dynamic microtubules.”, Curr Biol (Cover story), 2011.(*co-corresponding authors) doi: 10.1016/j.cub.2011.07.013
- Velve-Casquillas G, Fu C, Le Berre M, Cramer J, Meance S, Plecis A, Baigl D, Greffet JJ, Chen Y, Peil M, Tran PT“ Fast microfluidic temperature control for high resolution live cell imaging.”, Lab Chip, 2011. doi: 10.1039/c0lc00222d. 2011 Feb 7;11(3):484-9
- Zhang L, Shao H, Huang Y, Yan F, Chu Y, Hou H, Zhu M, Fu C, Aikhionbare F, Fang G, Ding X, Yao X“ Aikhionbare F, Fang G, Ding X, Yao X. 2011. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity.”, J Biol Chem, 2011. doi: 10.1074/jbc.M110.165340. 2011 Feb 7;11(3):484-9
- Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT.“Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation.”, Dev Cell, 2009. doi: 10.1016/j.devcel.2009.06.012
- Liu J, Wang Z, Jiang K, Zhang L, Zhao L, Hua S, Yan F, Yang Y, Wang D, Fu C, Ding X, Guo Z, Yao X“ PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis.”, J Biol Chem , 2011. doi: 10.1074/jbc.M109.009670. 2009 Aug 21;284(34):23059-71
- Fu C, Yan F, Wu F, Wu Q, Whittaker J, Hu H, Hu R, Yao X. “Mitotic phosphorylation of PRC1 at Thr470 is required for PRC1 oligomerization and proper central spindle organization.”, Cell Res, 2007. doi: 10.1038/cr.2007.32
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT“ Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast.”, Cell, 2011. doi: 10.1016/j.cell.2006.12.030. 2007 Jan 26;128(2):357-68
- Fang Z, Miao Y, Ding X, Deng H, Liu S, Wang F, Zhou R, Watson C, Fu C, Hu Q, Lillard JW, Jr., Powell M, Chen Y, Forte JG, Yao X“ Proteomic identification and functional characterization of a novel ARF6 GTPase-activating protein, ACAP4.”, Mol Cell Proteomics, 2006. doi: 10.1074/mcp.M600050-MCP200.2006 Aug;5(8):1437-49
- Xue Y, Zhou F, Fu C, Xu Y, Yao X“ SUMOsp: a web server for sumoylation site prediction.”, Nucleic Acids Res , 2006. doi: 10.1093/nar/gkl207. 2006 Jul 1;34(Web Server issue):W254-7
- Xue Y, Liu D, Fu C, Dou Z, Zhou Q, Yao X“ A novel genome-wide full-length kinesin prediction analysis reveals additional mammalian kinesins , Chinese Science Bulletin , 2006. doi: 51, 1836-1847
- Fu C, Ahmed K, Ding H, Ding X, Lan J, Yang Z, Miao Y, Zhu Y, Shi Y, Zhu J, Huang H, Yao X“ Stabilization of PML nuclear localization by conjugation and oligomerization of SUMO-3.”, Oncogene , 2005. doi: 10.1038/sj.onc.1208714. 2005 Aug 18;24(35):5401-13
- Yao J#, Fu C#, Ding X#, Guo Z, Zenreski A, Chen Y, Ahmed K, Liao J, Dou Z, Yao X“ Nek2A kinase regulates the localization of numatrin to centrosome in mitosis.”, FEBS Lett , 2004.(#equal contributions) doi: 10.1016/j.febslet.2004.08.047. 2004 Sep 24;575(1-3):112-8
- Zhou R, Watson C, Fu C,Yao X, Forte JG“ Myosin II is present in gastric parietal cells and required for lamellipodial dynamics associated with cell activation.”, Am J Physiol Cell Physiol , 2003. doi: 10.1152/ajpcell.00085.2003. 2003 Sep;285(3):C662-73
- Zhang J, Fu C, Miao Y, Dou Z, Yao X“ Protein kinase TTK interacts and co-localizes with CENP-E to the kinetochore of human cells.”, Chinese Science Bulletin , 2002.
2023
2022
2021
2020
2019
2018
2017
2016
2015
2014
2011
2009
2007
Before 2006